Impute numeric data below the threshold of measurement
Source:R/impute_lower.R
step_impute_lower.Rdstep_impute_lower() creates a specification of a recipe step designed for
cases where the non-negative numeric data cannot be measured below a known
value. In these cases, one method for imputing the data is to substitute the
truncated value by a random uniform number between zero and the truncation
point.
Usage
step_impute_lower(
recipe,
...,
role = NA,
trained = FALSE,
threshold = NULL,
skip = FALSE,
id = rand_id("impute_lower")
)Arguments
- recipe
A recipe object. The step will be added to the sequence of operations for this recipe.
- ...
One or more selector functions to choose variables for this step. See
selections()for more details.- role
Not used by this step since no new variables are created.
- trained
A logical to indicate if the quantities for preprocessing have been estimated.
- threshold
A named numeric vector of lower bounds. This is
NULLuntil computed byprep().- skip
A logical. Should the step be skipped when the recipe is baked by
bake()? While all operations are baked whenprep()is run, some operations may not be able to be conducted on new data (e.g. processing the outcome variable(s)). Care should be taken when usingskip = TRUEas it may affect the computations for subsequent operations.- id
A character string that is unique to this step to identify it.
Value
An updated version of recipe with the new step added to the
sequence of any existing operations.
Details
step_impute_lower() estimates the variable minimums from the data used in
the training argument of prep(). bake() then simulates a value for any
data at the minimum with a random uniform value between zero and the minimum.
As of recipes 0.1.16, this function name changed from step_lowerimpute()
to step_impute_lower().
Tidying
When you tidy() this step, a tibble is returned with
columns terms, value , and id:
- terms
character, the selectors or variables selected
- value
numeric, the estimated value
- id
character, id of this step
See also
Other imputation steps:
step_impute_bag(),
step_impute_knn(),
step_impute_linear(),
step_impute_mean(),
step_impute_median(),
step_impute_mode(),
step_impute_roll()
Examples
library(recipes)
data(biomass, package = "modeldata")
## Truncate some values to emulate what a lower limit of
## the measurement system might look like
biomass$carbon <- ifelse(biomass$carbon > 40, biomass$carbon, 40)
biomass$hydrogen <- ifelse(biomass$hydrogen > 5, biomass$carbon, 5)
biomass_tr <- biomass[biomass$dataset == "Training", ]
biomass_te <- biomass[biomass$dataset == "Testing", ]
rec <- recipe(
HHV ~ carbon + hydrogen + oxygen + nitrogen + sulfur,
data = biomass_tr
)
impute_rec <- rec |>
step_impute_lower(carbon, hydrogen)
tidy(impute_rec, number = 1)
#> # A tibble: 2 × 3
#> terms value id
#> <chr> <dbl> <chr>
#> 1 carbon NA impute_lower_b4CM3
#> 2 hydrogen NA impute_lower_b4CM3
impute_rec <- prep(impute_rec, training = biomass_tr)
tidy(impute_rec, number = 1)
#> # A tibble: 2 × 3
#> terms value id
#> <chr> <dbl> <chr>
#> 1 carbon 40 impute_lower_b4CM3
#> 2 hydrogen 5 impute_lower_b4CM3
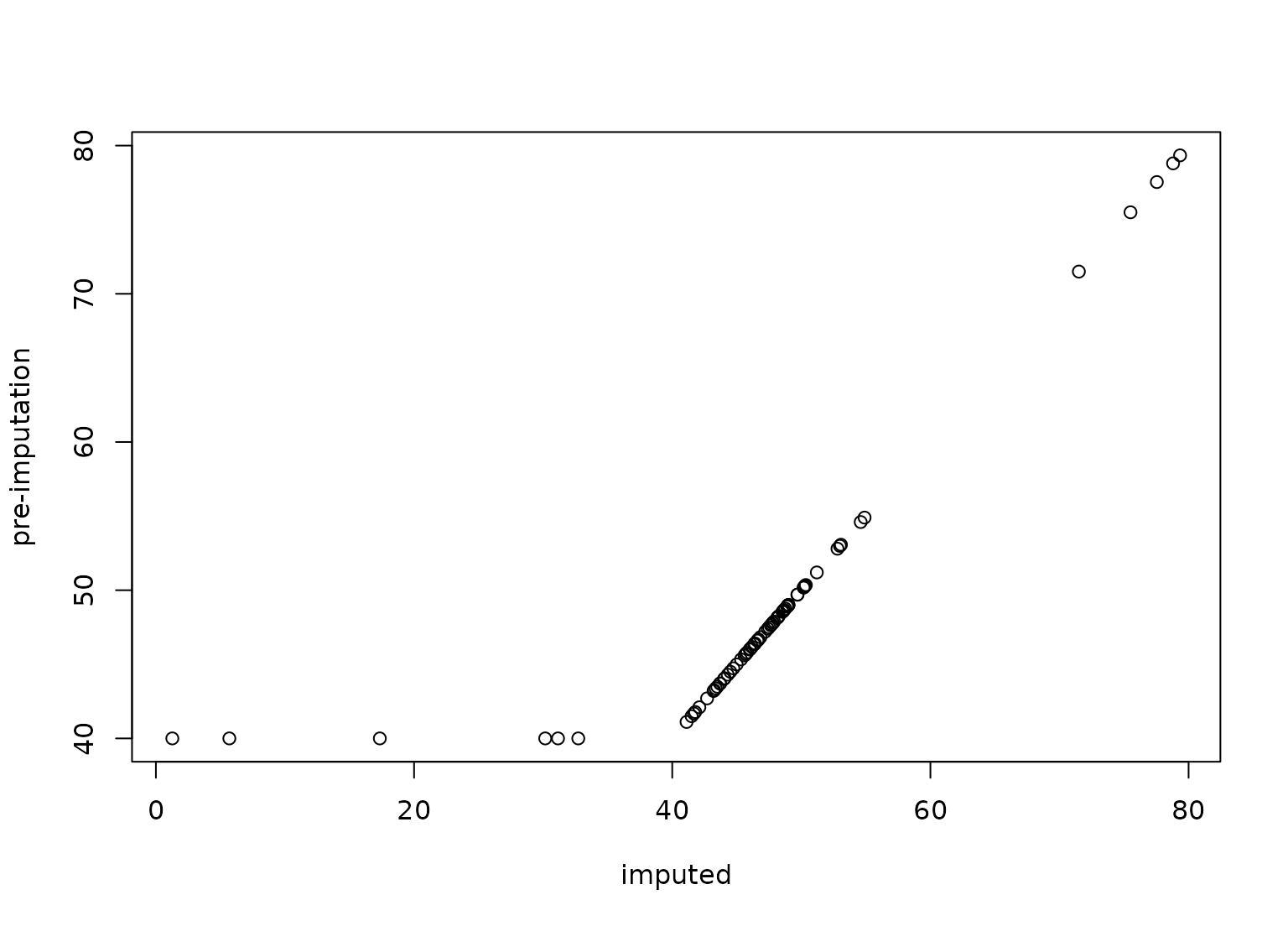
transformed_te <- bake(impute_rec, biomass_te)
plot(transformed_te$carbon, biomass_te$carbon,
ylab = "pre-imputation", xlab = "imputed"
)