step_spatialsign() is a specification of a recipe step that will convert
numeric data into a projection on to a unit sphere.
Usage
step_spatialsign(
recipe,
...,
role = "predictor",
na_rm = TRUE,
trained = FALSE,
columns = NULL,
skip = FALSE,
id = rand_id("spatialsign")
)Arguments
- recipe
A recipe object. The step will be added to the sequence of operations for this recipe.
- ...
One or more selector functions to choose variables for this step. See
selections()for more details.- role
For model terms created by this step, what analysis role should they be assigned? By default, the new columns created by this step from the original variables will be used as predictors in a model.
- na_rm
A logical: should missing data be removed from the norm computation?
- trained
A logical to indicate if the quantities for preprocessing have been estimated.
- columns
A character string of the selected variable names. This field is a placeholder and will be populated once
prep()is used.- skip
A logical. Should the step be skipped when the recipe is baked by
bake()? While all operations are baked whenprep()is run, some operations may not be able to be conducted on new data (e.g. processing the outcome variable(s)). Care should be taken when usingskip = TRUEas it may affect the computations for subsequent operations.- id
A character string that is unique to this step to identify it.
Value
An updated version of recipe with the new step added to the
sequence of any existing operations.
Details
The spatial sign transformation projects the variables onto a unit sphere and
is related to global contrast normalization. The spatial sign of a vector w
is w/norm(w).
The variables should be centered and scaled prior to the computations.
Tidying
When you tidy() this step, a tibble is returned with
columns terms and id:
- terms
character, the selectors or variables selected
- id
character, id of this step
Case weights
This step performs an unsupervised operation that can utilize case weights.
As a result, only frequency weights are allowed. For more information, see
the documentation in case_weights and the examples on tidymodels.org.
Unlike most, this step requires the case weights to be available when new
samples are processed (e.g., when bake() is used or predict() with a
workflow). To tell recipes that the case weights are required at bake time,
use recipe |> update_role_requirements(role = "case_weights", bake = TRUE). See update_role_requirements() for more information.
References
Serneels, S., De Nolf, E., and Van Espen, P. (2006). Spatial sign preprocessing: a simple way to impart moderate robustness to multivariate estimators. Journal of Chemical Information and Modeling, 46(3), 1402-1409.
See also
Other multivariate transformation steps:
step_classdist(),
step_classdist_shrunken(),
step_depth(),
step_geodist(),
step_ica(),
step_isomap(),
step_kpca(),
step_kpca_poly(),
step_kpca_rbf(),
step_mutate_at(),
step_nnmf(),
step_nnmf_sparse(),
step_pca(),
step_pls(),
step_ratio()
Examples
data(biomass, package = "modeldata")
biomass_tr <- biomass[biomass$dataset == "Training", ]
biomass_te <- biomass[biomass$dataset == "Testing", ]
rec <- recipe(
HHV ~ carbon + hydrogen + oxygen + nitrogen + sulfur,
data = biomass_tr
)
ss_trans <- rec |>
step_center(carbon, hydrogen) |>
step_scale(carbon, hydrogen) |>
step_spatialsign(carbon, hydrogen)
ss_obj <- prep(ss_trans, training = biomass_tr)
transformed_te <- bake(ss_obj, biomass_te)
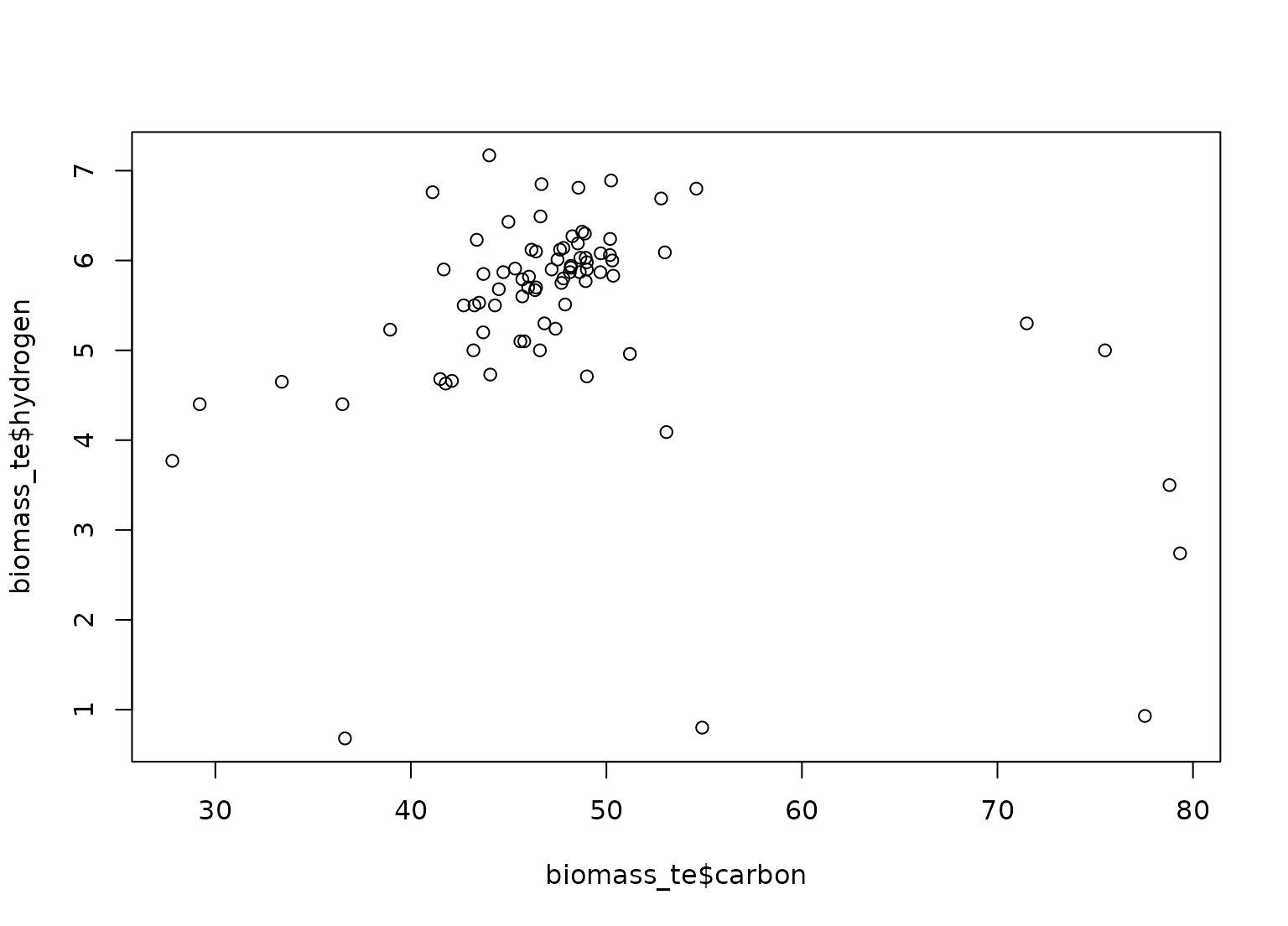
plot(biomass_te$carbon, biomass_te$hydrogen)
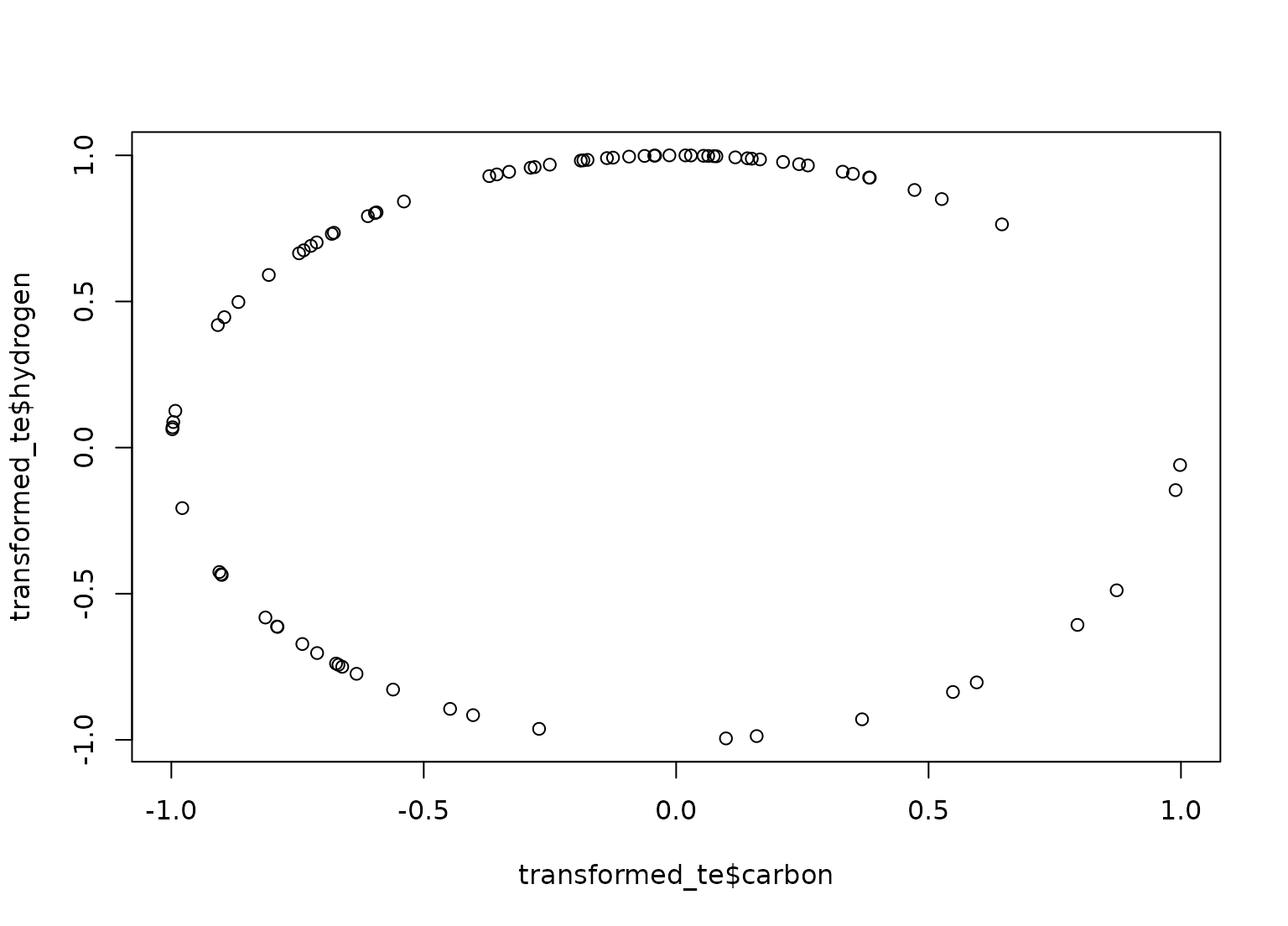
 plot(transformed_te$carbon, transformed_te$hydrogen)
plot(transformed_te$carbon, transformed_te$hydrogen)
 tidy(ss_trans, number = 3)
#> # A tibble: 2 × 2
#> terms id
#> <chr> <chr>
#> 1 carbon spatialsign_Geidk
#> 2 hydrogen spatialsign_Geidk
tidy(ss_obj, number = 3)
#> # A tibble: 2 × 2
#> terms id
#> <chr> <chr>
#> 1 carbon spatialsign_Geidk
#> 2 hydrogen spatialsign_Geidk
tidy(ss_trans, number = 3)
#> # A tibble: 2 × 2
#> terms id
#> <chr> <chr>
#> 1 carbon spatialsign_Geidk
#> 2 hydrogen spatialsign_Geidk
tidy(ss_obj, number = 3)
#> # A tibble: 2 × 2
#> terms id
#> <chr> <chr>
#> 1 carbon spatialsign_Geidk
#> 2 hydrogen spatialsign_Geidk
